Data Inspection- Preprocessing - Unsupervised ML#
This tutorial demonstrates the most important basic steps involved in the analysis of scanning electron diffraction data.
The data was acquired from a GaAs nanowire adopting the zinc blende structure and exhibiting type I twinning (i.e. on {111}) along its length. The region of interest contains a single nanowire comprising multiple crystals each in one of the two twinned orientations and near to a <1-10> zone axis.
This functionaility has been checked to run in pyxem version 0.14.1 (April 2022). However, bugs are always possible, do not trust the code blindly, and if you experience any issues please report them here: pyxem/pyxem-demos#issues
Contents#
Loading & Inspection
Alignment & Calibration
Virtual Diffraction Imaging
Machine Learning SPED Data
Peak Finding
Import pyxem and other required libraries
[ ]:
# Changing the matplotlib background will give you interactive
#%matplotlib qt5
[ ]:
%matplotlib inline
import hyperspy.api as hs
import pyxem as pxm
import numpy as np
WARNING:silx.opencl.common:Unable to import pyOpenCl. Please install it from: https://pypi.org/project/pyopencl
1. Loading and Inspection#
Load the SPED data acquired from the nanowire
[ ]:
dp = hs.load('./data/01/twinned_nanowire.hdf5', reader="hspy")
/Users/carterfrancis/mambaforge/envs/pyxem-demos/lib/python3.11/site-packages/hyperspy/misc/utils.py:471: VisibleDeprecationWarning: Use of the `binned` attribute in metadata is going to be deprecated in v2.0. Set the `axis.is_binned` attribute instead.
warnings.warn(
/Users/carterfrancis/mambaforge/envs/pyxem-demos/lib/python3.11/site-packages/hyperspy/io.py:572: VisibleDeprecationWarning: Loading old file version. The binned attribute has been moved from metadata.Signal to axis.is_binned. Setting this attribute for all signal axes instead.
warnings.warn('Loading old file version. The binned attribute '
Inspect the dp object
[ ]:
dp
<Diffraction2D, title: , dimensions: (30, 100|144, 144)>
Specify that the data is electron diffraction data
[ ]:
dp.set_signal_type('electron_diffraction')
Inspect the signal type
[ ]:
dp
<ElectronDiffraction2D, title: , dimensions: (30, 100|144, 144)>
Inspect the data type of the object
[ ]:
dp.data.dtype
dtype('uint8')
Inspect the metadata associated with the object ‘dp’
[ ]:
dp.metadata
- Acquisition_instrument
- TEM
- beam_energy = 300.0
- camera_length = 0.21000000000000002
- scan_rotation = 277.0
- General
- FileIO
- 0
- hyperspy_version = 1.7.5
- io_plugin = hyperspy.io_plugins.hspy
- operation = load
- timestamp = 2023-10-26T10:27:31.436805-05:00
- original_filename = nanowire_precession.blo
- time = (2014, 12, 8)
- title =
- Signal
- signal_origin =
- signal_type = electron_diffraction
Set important experimental parameters using the built in function
[ ]:
dp.set_experimental_parameters(beam_energy=300.0,
camera_length=21.0,
scan_rotation=277.0,
convergence_angle=0.7,
exposure_time=10.0)
See how this changed the metadata
[ ]:
dp.metadata
- Acquisition_instrument
- TEM
- Detector
- Diffraction
- camera_length = 21.0
- exposure_time = 10.0
- beam_energy = 300.0
- camera_length = 0.21000000000000002
- convergence_angle = 0.7
- scan_rotation = 277.0
- General
- FileIO
- 0
- hyperspy_version = 1.7.5
- io_plugin = hyperspy.io_plugins.hspy
- operation = load
- timestamp = 2023-10-26T10:27:31.436805-05:00
- original_filename = nanowire_precession.blo
- time = (2014, 12, 8)
- title =
- Signal
- signal_origin =
- signal_type = electron_diffraction
Set another metadata item and check it
[ ]:
dp.metadata.set_item("General.title", 'GaAs Nanowire')
dp.metadata
- Acquisition_instrument
- TEM
- Detector
- Diffraction
- camera_length = 21.0
- exposure_time = 10.0
- beam_energy = 300.0
- camera_length = 0.21000000000000002
- convergence_angle = 0.7
- scan_rotation = 277.0
- General
- FileIO
- 0
- hyperspy_version = 1.7.5
- io_plugin = hyperspy.io_plugins.hspy
- operation = load
- timestamp = 2023-10-26T10:27:31.436805-05:00
- original_filename = nanowire_precession.blo
- time = (2014, 12, 8)
- title = GaAs Nanowire
- Signal
- signal_origin =
- signal_type = electron_diffraction
[ ]:
dp
<ElectronDiffraction2D, title: GaAs Nanowire, dimensions: (30, 100|144, 144)>
Plot the data to inspect it
[ ]:
dp.plot(cmap='inferno', vmax=50)
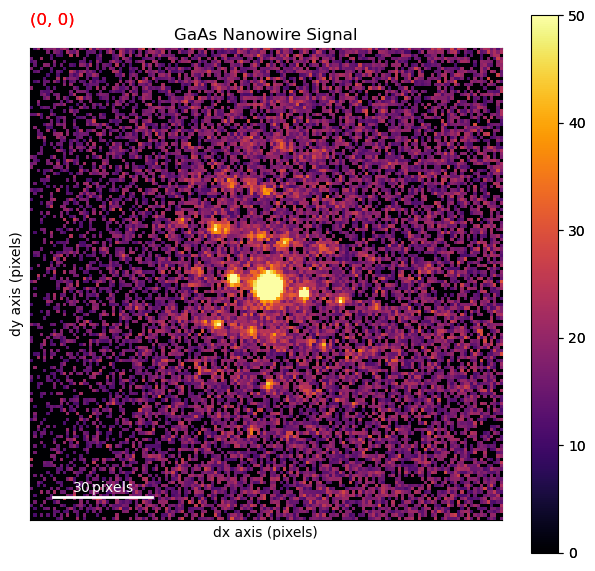
2. Alignment & Calibration#
Apply distortion corrections to the data due to off-axis acquisition
[ ]:
dp.apply_affine_transformation(np.array([[0.99,0,0],
[0,0.69,0],
[0,0,1]]),
keep_dtype=True)
[########################################] | 100% Completed | 2.50 ss
[ ]:
dp.data.dtype
dtype('uint8')
Align the dataset based on the direct beam position
[ ]:
dp.center_direct_beam(method='cross_correlate',
radius_start=2,
radius_finish=5,
half_square_width=10)
[########################################] | 100% Completed | 32.64 s
[########################################] | 100% Completed | 1.99 ss
[ ]:
dp.plot(cmap='inferno', vmax=50)
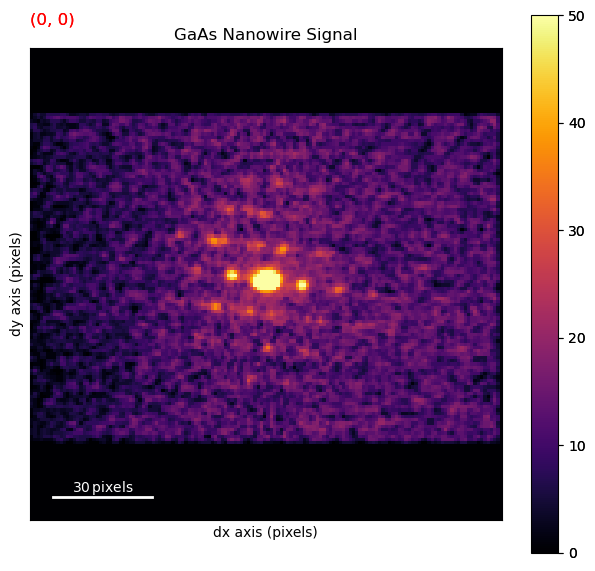
Measure known interplanar spacing to obtain calibration
[ ]:
dpm = dp.mean((0,1))
dpm.plot(cmap='inferno', vmax=50)
line = hs.roi.Line2DROI(x1=25.8525, y1=64.5691, x2=120.907, y2=77.0079, linewidth=5.49734)
line.add_widget(dpm)
<hyperspy.drawing._widgets.line2d.Line2DWidget at 0x1574b0290>
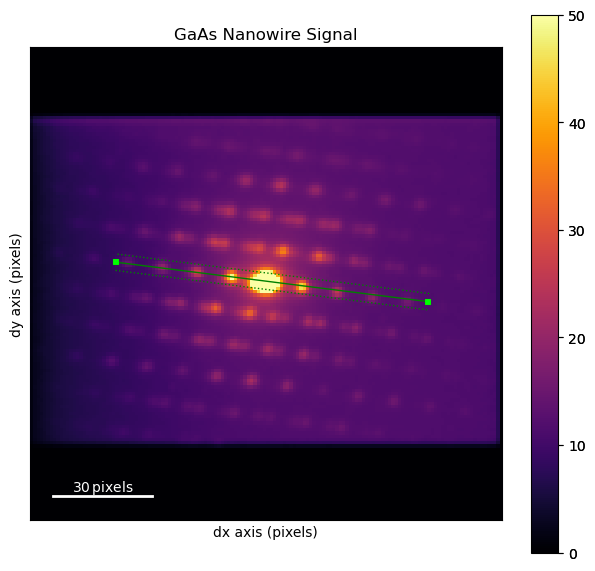
[ ]:
trace = line(dpm)
trace.plot()
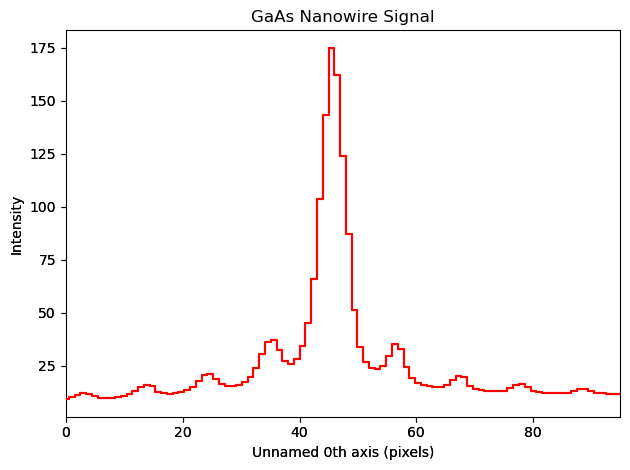
[ ]:
recip_d111 = np.sqrt((3/5.6535**2))
recip_cal = recip_d111 / 11.4
Set data calibrations
[ ]:
dp.set_diffraction_calibration(recip_cal)
dp.set_scan_calibration(10)
Plot the calibrated data
[ ]:
dp.plot(cmap='inferno', vmax=50)
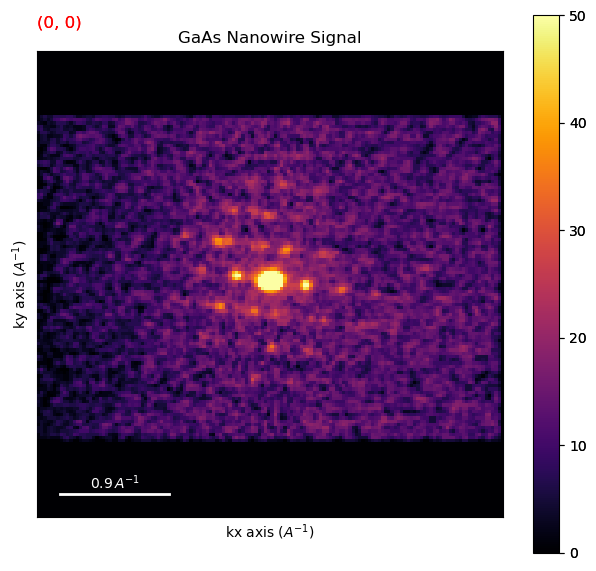
3. Virtual Diffraction Imaging & Selecting Regions#
3.1 Interactive VDF Imaging#
Plot an interactive virtual image integrating intensity within a circular subset of pixels in the diffraction pattern
[ ]:
roi = hs.roi.CircleROI(cx=0.,cy=0, r_inner=0, r=0.07)
[ ]:
dp.plot_integrated_intensity(roi)
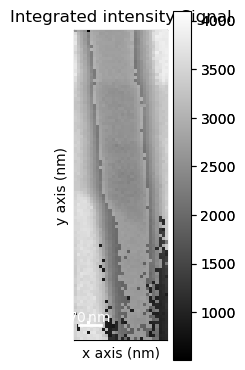
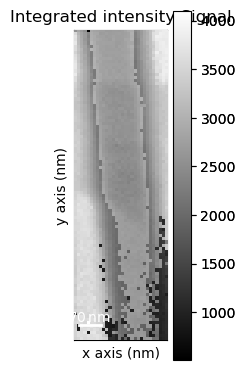
Get the virtual diffraction image associated with the last integration window used interactively
[ ]:
vdf = dp.get_integrated_intensity(roi)
Plot the virtual dark-field image
[ ]:
vdf.plot()
Inspect the metadata
[ ]:
vdf.metadata
- Acquisition_instrument
- TEM
- Detector
- Diffraction
- camera_length = 21.0
- exposure_time = 10.0
- beam_energy = 300.0
- camera_length = 0.21000000000000002
- convergence_angle = 0.7
- scan_rotation = 277.0
- Diffraction
- integrated_range = CircleROI(cx=0, cy=0, r=0.07, r_inner=0) of GaAs Nanowire
- General
- FileIO
- 0
- hyperspy_version = 1.7.5
- io_plugin = hyperspy.io_plugins.hspy
- operation = load
- timestamp = 2023-10-26T10:27:31.436805-05:00
- original_filename = nanowire_precession.blo
- time = (2014, 12, 8)
- title = Integrated intensity
- Signal
- signal_origin =
- signal_type =
Save the virtual dark-field image as a 32bit tif
[ ]:
vdf.change_dtype('float32')
vdf.save('vdfeg.tif')
3.2 Form multiple images using a VDF Generator#
Import the VirtualImageGenerator class
[ ]:
from pyxem.generators.virtual_image_generator import VirtualImageGenerator
Initialize the VDFGenerator
[ ]:
vdfgen = VirtualImageGenerator(dp)
Calculate 10 annular VDF images between 0 and 1 reciprocal angstroms
[ ]:
vdfs = vdfgen.get_concentric_virtual_images(k_min=0,
k_max=1,
k_steps=10)
Plot the VDF images
[ ]:
vdfs.plot()
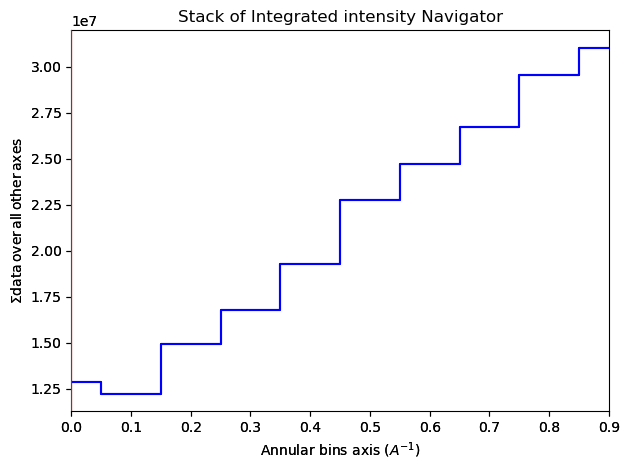
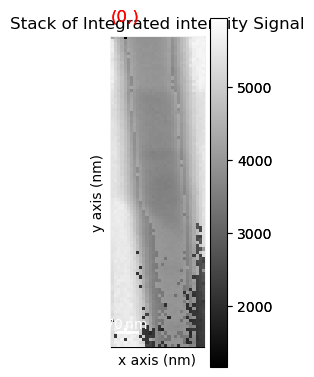
Save the stack of VDF image as a 32bit tif stack
[ ]:
vdfs.change_dtype('float32')
vdfs.save('vdsfeg.tif')
3.3 Select a region in the scan#
Plot the data with an adjustable marker indicating where to crop the scan region
[ ]:
reg = hs.roi.RectangularROI(left=50, top=630, right=290, bottom=870)
dp.plot(cmap='inferno', vmax=50)
reg.add_widget(dp)
<hyperspy.drawing._widgets.rectangles.RectangleWidget at 0x157384150>
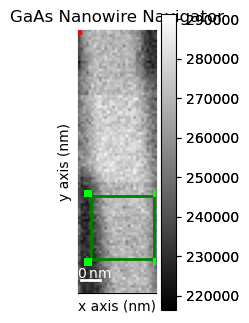
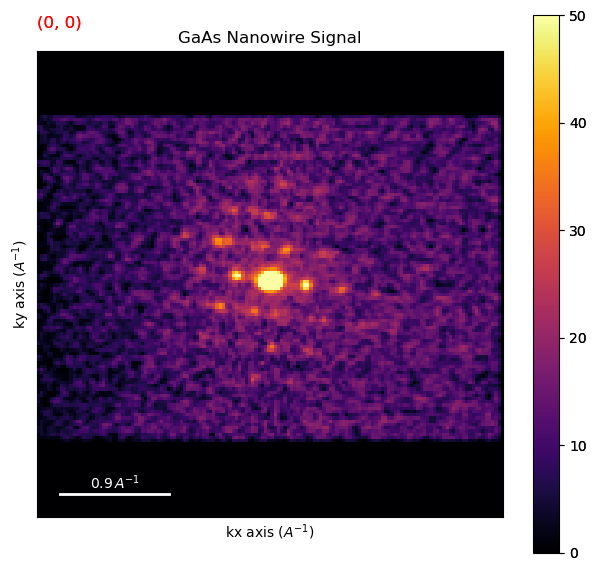
Crop the dataset based on the region defined above
[ ]:
dpc = reg(dp)
Calculate the mean diffraction pattern from the selected region
[ ]:
dpcm = dpc.mean((0,1))
Plot the mean diffraction pattern from the selected region
[ ]:
dpcm.plot(cmap='inferno', vmax=50)
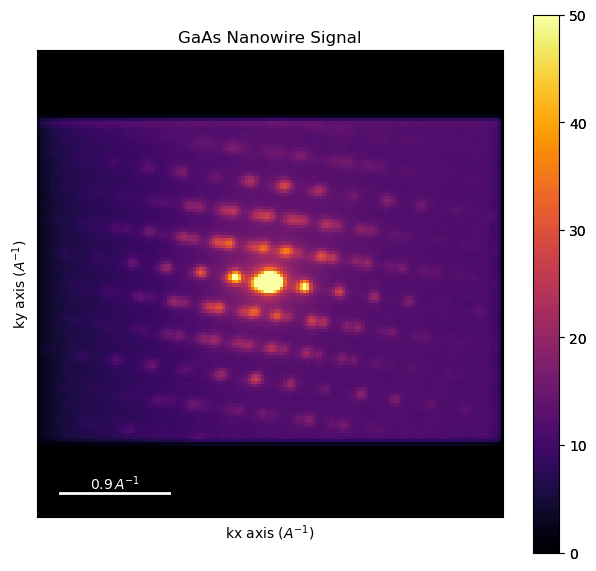
4. Unsupervised learning#
Perform singular value decomposition (SVD) of the data
[ ]:
dpc.data =dpc.data+1 # Sometimes the SVD Solver won't converge if there are a lot of zeros.
[ ]:
dpc.data = dpc.data.astype('float64')
dpc.decomposition(True, algorithm='SVD')
Decomposition info:
normalize_poissonian_noise=True
algorithm=SVD
output_dimension=None
centre=None
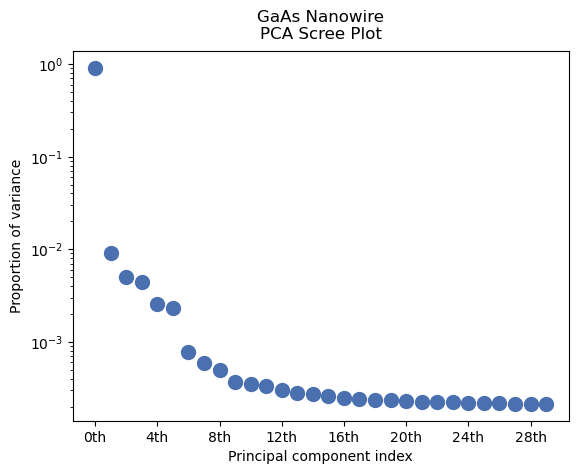
Obtain a “Scree plot” by plotting the fraction of variance described by each principal component
[ ]:
dpc.plot_explained_variance_ratio()
<Axes: title={'center': 'GaAs Nanowire\nPCA Scree Plot'}, xlabel='Principal component index', ylabel='Proportion of variance'>
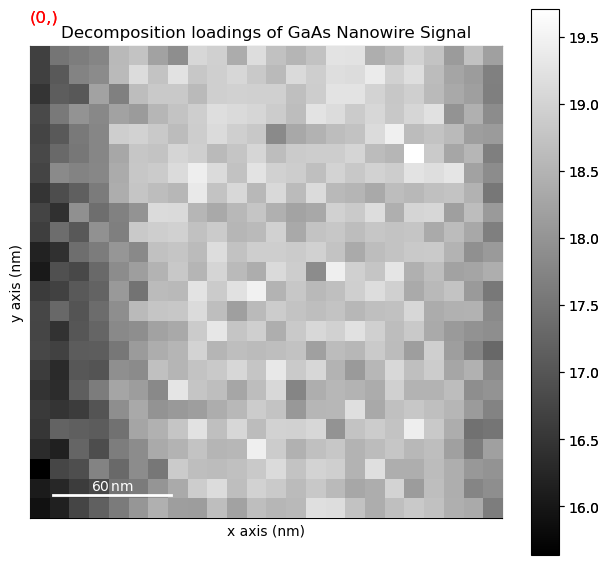
Plot the decomposition results and have a look at them
[ ]:
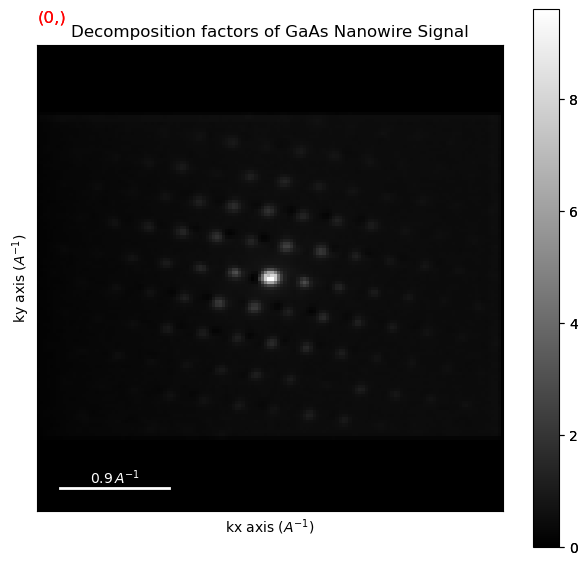
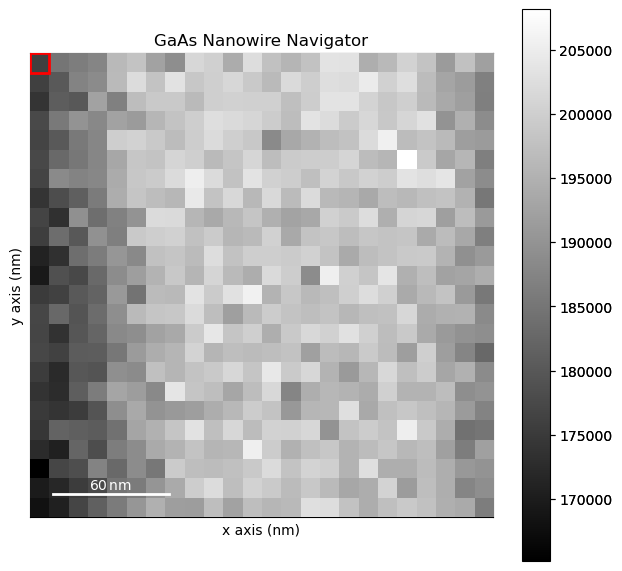
dpc.plot_decomposition_results()
WARNING:hyperspy.drawing.mpl_he:Navigation sliders not available. No toolkit registered. Install hyperspy_gui_ipywidgets or hyperspy_gui_traitsui GUI elements.
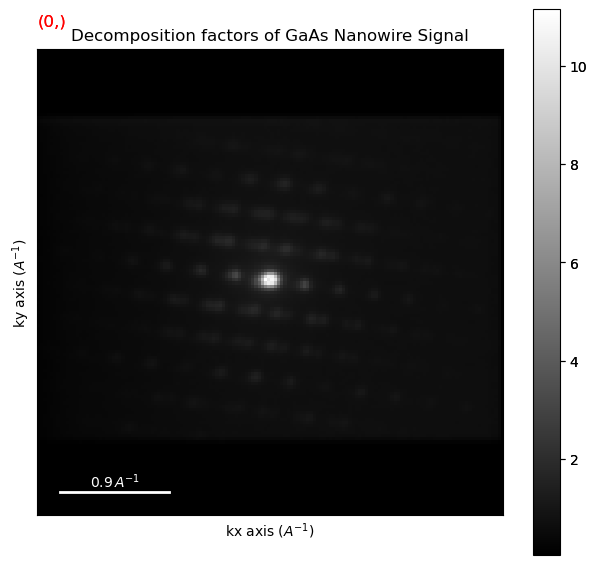
Perform non-negative matrix factorisation (NMF)
[ ]:
dpc.decomposition(True, algorithm='NMF', output_dimension=4,)
Decomposition info:
normalize_poissonian_noise=True
algorithm=NMF
output_dimension=4
centre=None
scikit-learn estimator:
NMF(n_components=4)
/Users/carterfrancis/mambaforge/envs/pyxem-demos/lib/python3.11/site-packages/sklearn/decomposition/_nmf.py:1710: ConvergenceWarning: Maximum number of iterations 200 reached. Increase it to improve convergence.
warnings.warn(
Plot the NMF results
[ ]:
dpc.plot_decomposition_results()
WARNING:hyperspy.drawing.mpl_he:Navigation sliders not available. No toolkit registered. Install hyperspy_gui_ipywidgets or hyperspy_gui_traitsui GUI elements.
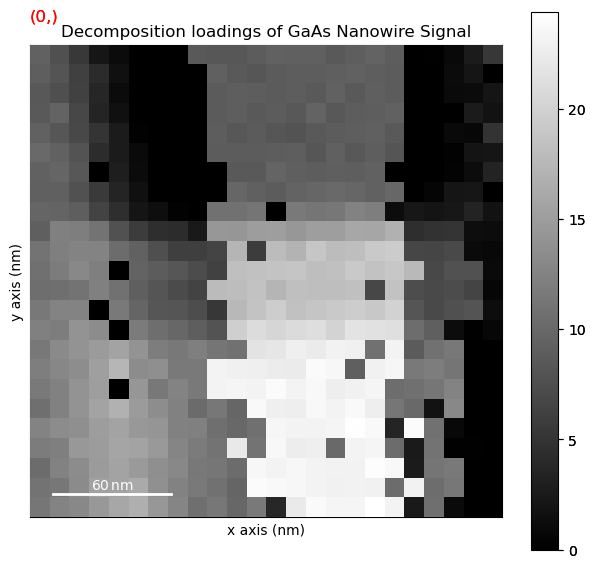
5. Peak Finding#
Perform peak finding on all diffraction patterns in data
[ ]:
peaks = dpc.find_peaks(method='difference_of_gaussian',
min_sigma=1.,
max_sigma=6.,
sigma_ratio=1.6,
threshold=0.04,
overlap=0.99,
interactive=False)
[########################################] | 100% Completed | 3.47 ss
Check the peaks object type
[ ]:
peaks
<BaseSignal, title: GaAs Nanowire, dimensions: (24, 24|ragged)>
Look at what’s in the peaks object
[array([[60, 58],
[70, 61],
[61, 69],
[71, 72],
[82, 73],
[73, 82],
[83, 84]])]
[array([[59, 58],
[70, 61],
[61, 69],
[71, 71],
[82, 73],
[73, 82],
[83, 84]])]
coaxing peaks back into a DiffractionVectors
[ ]:
from pyxem.signals.diffraction_vectors import DiffractionVectors
[ ]:
peaks = DiffractionVectors.from_peaks(peaks,center=(72,72),calibration=recip_cal)
WARNING:hyperspy.signal:The function you applied does not take into account the difference of units and of scales in-between axes.
[########################################] | 100% Completed | 105.83 ms
Plot found peak positions as an overlay on the data
[ ]:
peaks.plot_diffraction_vectors_on_signal(dpc, cmap='gray', vmax=50)
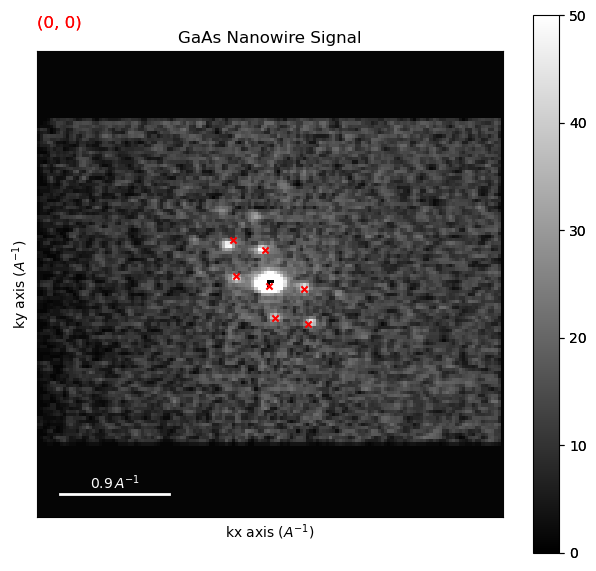
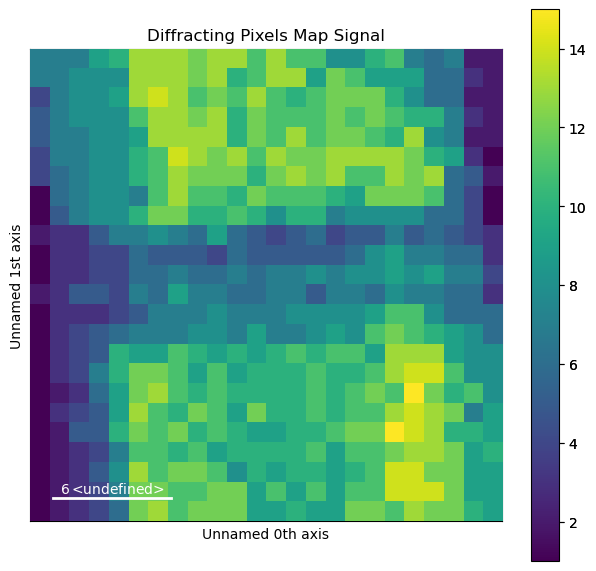
Form a diffracting pixels map to show where peaks were found and plot it
[ ]:
crystim = peaks.get_diffracting_pixels_map(binary=False)
crystim.plot(cmap='viridis')
WARNING:hyperspy.signal:The function you applied does not take into account the difference of units and of scales in-between axes.
WARNING:hyperspy.io:`signal_type='diffraction_vectors'` not understood. See `hs.print_known_signal_types()` for a list of installed signal types or https://github.com/hyperspy/hyperspy-extensions-list for the list of all hyperspy extensions providing signals.
[########################################] | 100% Completed | 106.79 ms
WARNING:hyperspy.io:`signal_type='signal2d'` not understood. See `hs.print_known_signal_types()` for a list of installed signal types or https://github.com/hyperspy/hyperspy-extensions-list for the list of all hyperspy extensions providing signals.